Bee DNA Barcoding at Pima Community College (PCC)- a review in pictures
Students in “General Biology for Majors I” (Bio 181) classes at the Northwest Campus participate in a Course-based Undergraduate Research Experience (CURE). PCC students identify wild bee species by extracting and analyzing DNA through a process called DNA barcoding. Students provide bee species identifications to researchers at the Arizona Sonora Desert Museum and the University of Arizona. Researchers must first identify species in order to better monitor and conserve these important pollinators. The Sonoran Desert is likely to have the highest bee diversity worldwide but this has not been well studied.
The students performed the extraction and analysis with help from the our University of Arizona (UA) Department of Entomology partners, in particular the Insect Collection. They discussed the results with our Arizona Sonora Desert Museum partners. They did an end of semester poster session. Several students traveled to Washington D.C. for the 2023 National Science Foundation’s Emerging Researchers Network Conference to present their results and attend workshops on educational opportunities and STEM careers.
Student Posters and DNA Barcode Results
PCC Bio181 General Bio Fall 2020 AM
PCC General Biology Bio181 Fall 2020 PM
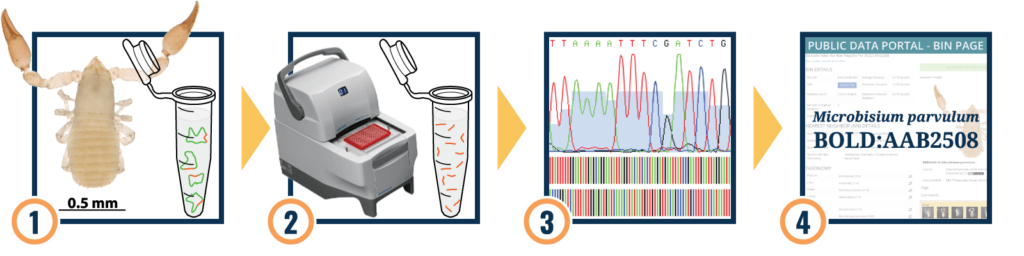
1. DNA Extraction
2. Polymerase Chain Reaction (PCR)–make copies of the DNA of Interest
3. Send samples to commercial lab for DNA sequencing
4. View results of sequencing using DNA Subway and Barcode of Life Data System (Bold)
1. DNA Extraction
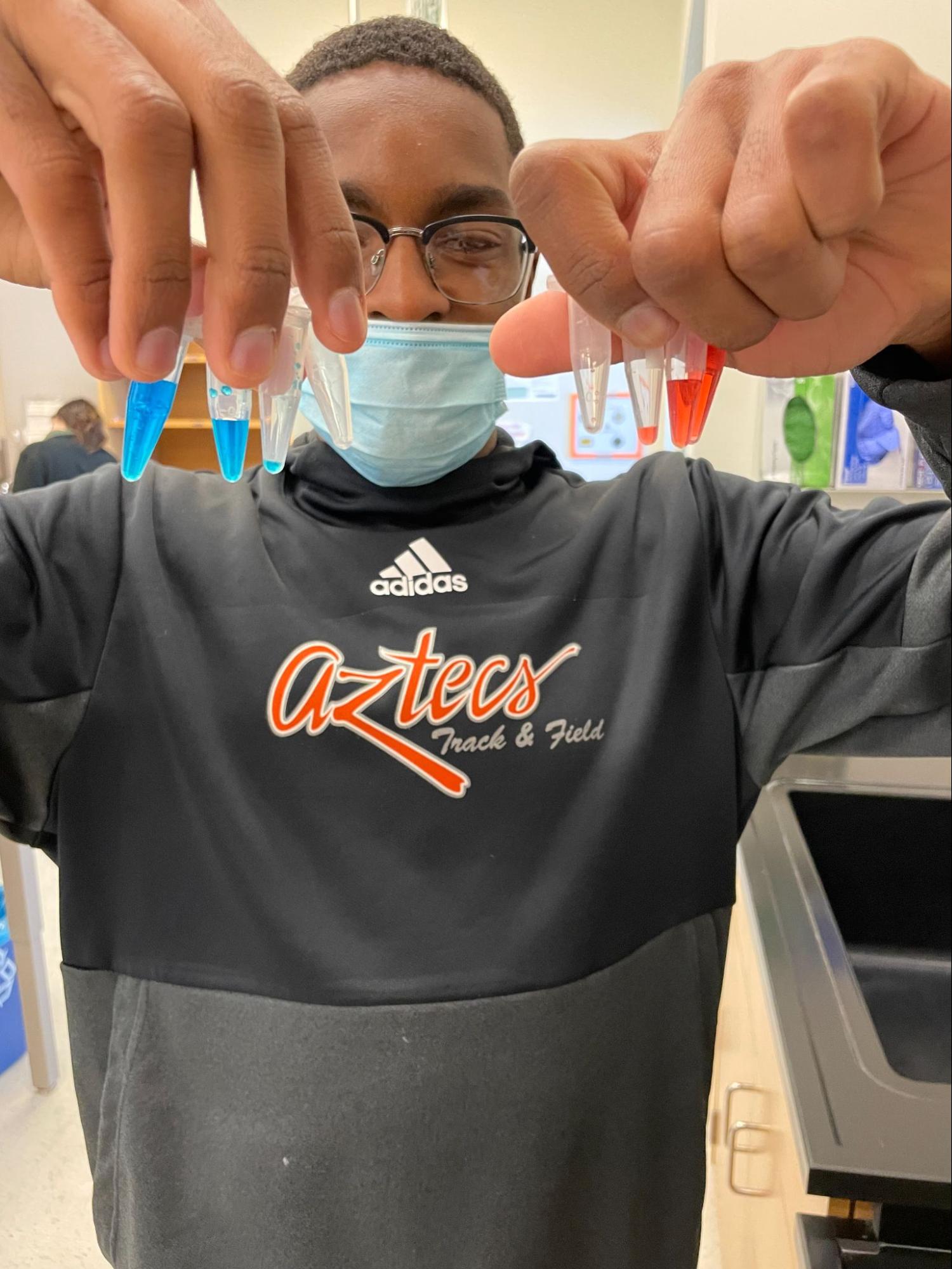


Back to top
2. Polymerase Chain Reaction (PCR) – make many copies of the DNA sequence of interest


Back to top
3. Samples sent out for DNA sequencing (no photos)
Back to top
4. View results of DNA sequencing using DNA Subway and Barcode of Life Database (BOLD)

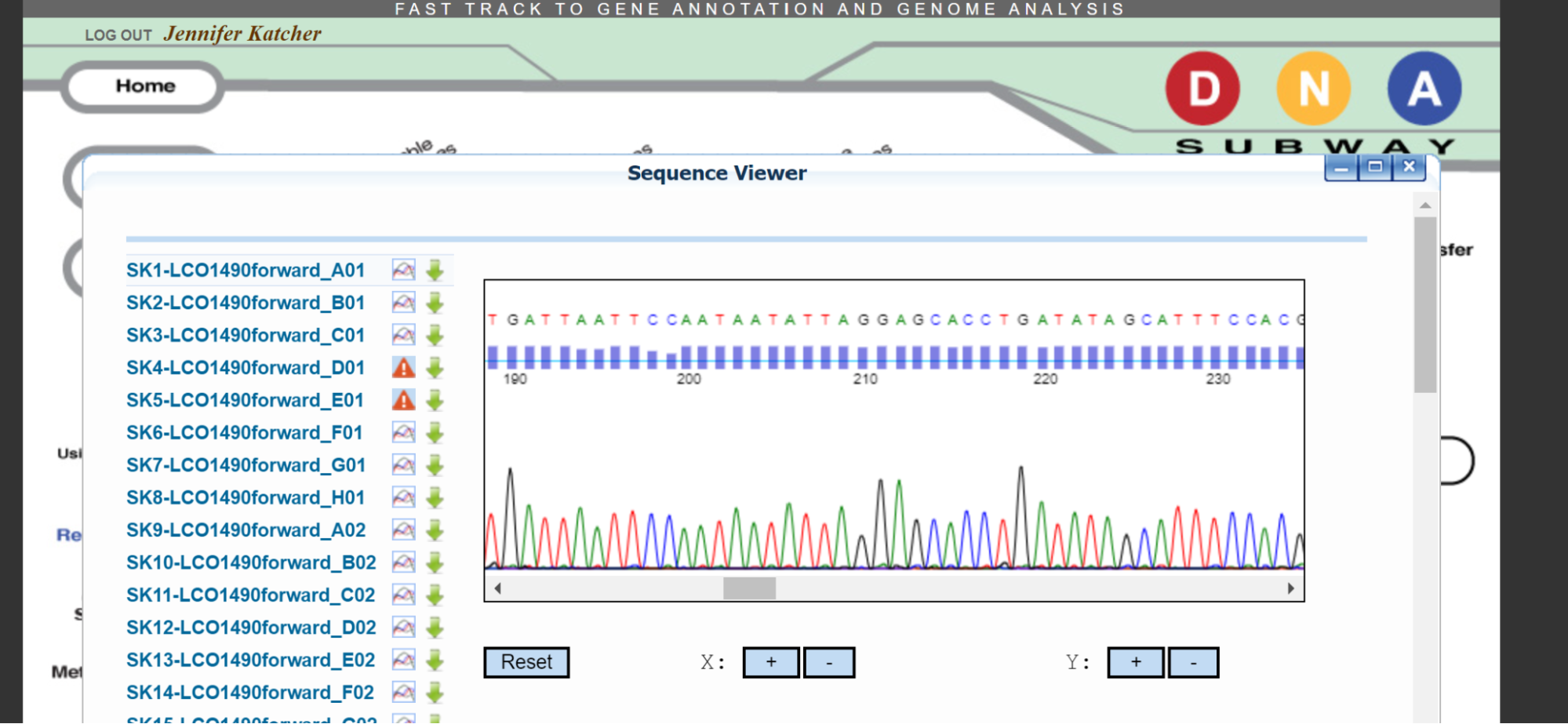
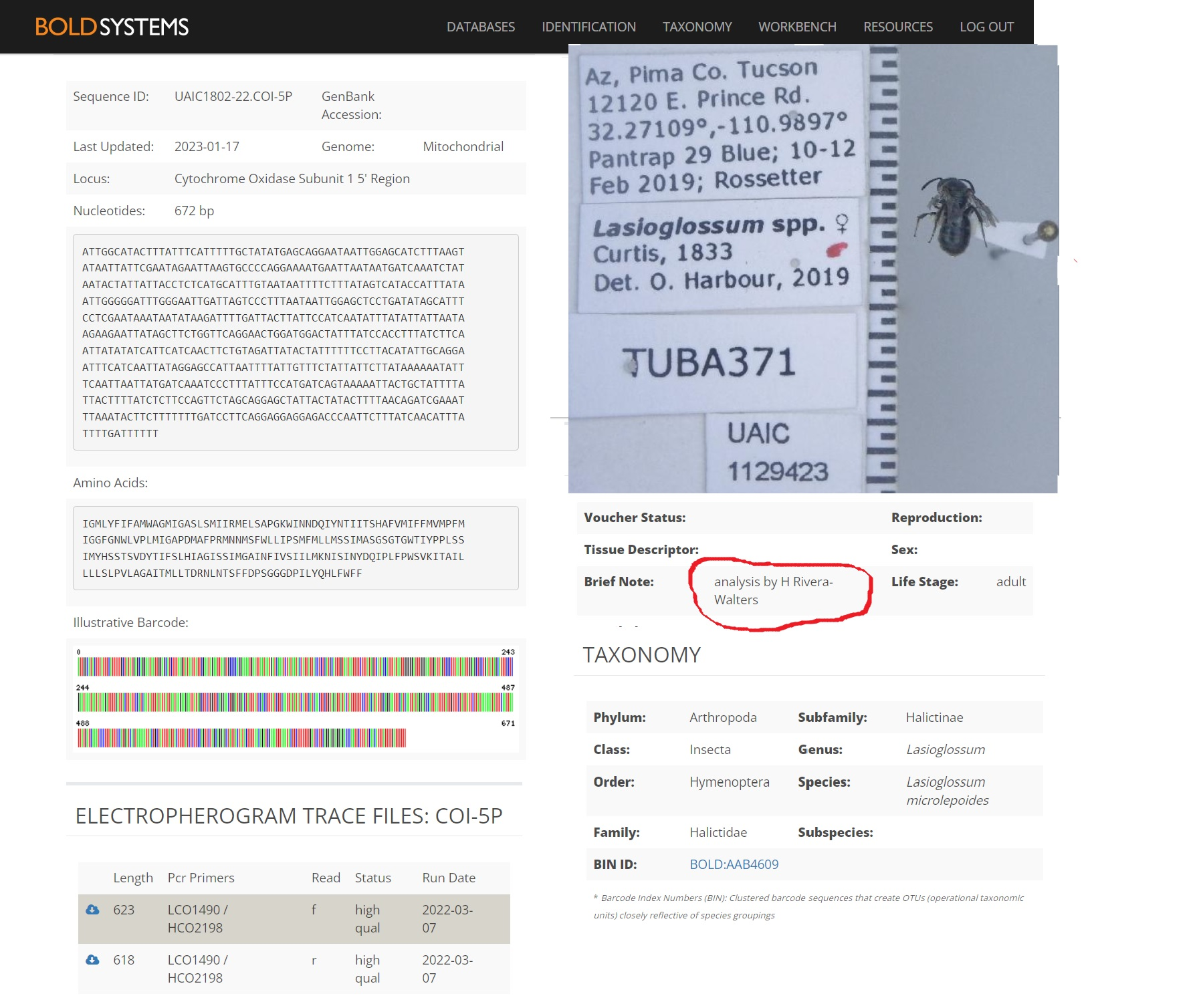
Here’s a link to the students’ records published in the the Barcode of Life Data System.
Back to top
Results Discussion
Kim Franklin, our Arizona Sonora Desert Museum research partner, met with students to discuss their results. Following Kim’s visit, students wrote a paragraph summarizing how their work supports the Tucson Bee Collaborative. Here’s a paragraph from a student:
Our class relates to the activities of Tucson Bee Collaborative through our mutual interest in cells down to the DNA. Our collaboration allows us the students to understand things by applying them in the real world, the tremendous benefit, experience and honor would have otherwise been lost. It is also productive and helpful to the Tucson Bee Collaborative, since it gives them data quicker and cheaper. Not only do we share mutual benefits, we also share the love of science and the attempt to leave a positive contribution and have our names remembered in the history of science. They are interested in the identification of the bee’s species, and we are interested in learning more and more science. This is a great collaboration that I am grateful for, and nothing is greater than a partnership that results in good for the community.
“Nothing is greater than a partnership that results in good for the community”–We couldn’t agree more!
Public Outreach and Science Engagement:
Students communicated their results to interested community members at an end-of semester poster session and demonstration.



Six students traveled to Washington DC on Feb. 9-12, 2023 to attend the National Science Foundation’s Emerging Researchers Network conference. They presented research results from their bee DNA analysis work. Students also attended workshops to learn about future educational opportunities and STEM careers.

Educational research supported by NSF HSI program Award #1928400
Bee DNA Barcoding CURE Educational Research Outcomes (from 2022 NSF annual report)–
- Students’ research self-efficacy increased throughout the course.
- Students’ science identity increased throughout the course.
- The course increased students’ knowledge of how to engage with their community, and for some students, increased their self-efficacy in doing so.
PCC DNA Barcoding CURE by the numbers–
- 316 students have participated in TBC DNA barcoding (F17- current)
- 204 students have visited the UA and toured the UAIC (F17 – F19, S23)
- 150 students have/will create and present posters in person (F21 – S23)
- 46 students have created and presented posters virtually (F20)
- 198 students have published records in the peer-reviewed Barcode of Life Database (BOLD) (F17-S22).
- These records include 151 DNA sequences representing 85 different species.
